Titin is the largest known protein, at around 3.5 MDa (the etymological link to Titan is intentional). It acts as a molecular spring in muscle and is a prime target for protein unfolding studies. Previously, direct comparison between molecular simulations and single-molecule unfolding experiments was not possible because of the huge difference in pulling velocity exerted on the molecule. Now, researchers in the Scheuring Lab at Université Aix-Marseille have developed a high-speed force spectroscopy (HS-FS) method that stretches titin molecules at speeds more akin to simulations. Lab director Simon Scheuring answers questions about the technique.
The technique is atomic force microscopy (AFM)-based force spectroscopy. The protein molecule is tethered between the AFM tip (at the end of the AFM cantilever) and a solid support that is pulled backwards by a piezoelectric element. When this is done, the protein molecule is stretched and unfolded. The AFM cantilever reports the forces required for this process (See Figure 1).
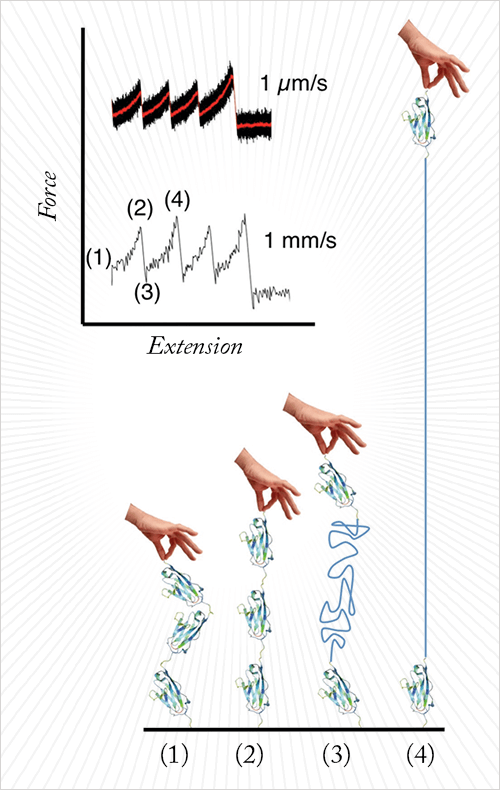
Compared to conventional AFM force spectroscopy, we used cantilevers that are about 30 times shorter than conventional cantilevers (6 µm versus 200 µm). We also used a novel sample support to minimize hydrodynamic drag and allow very fast electronics. Each of these ameliorations was essential to pull about 1000 times faster than a conventional setup.
We read the data (the cantilever deflection) out at frequency of 2MHz, or two million data points per second. When we unfold a protein that is 25 nm in length at a speed of 4 millimetres per second, we only retrieve about 15 data points. Therefore, to unfold molecules even faster, we must further improve our electronics. As you can imagine, we also acquire enormous amounts of data during a long experiment.
Simulations have become very important in modern biology (see “The New Model”). In silico simulations provide full-atomistic movies of what molecules are ‘doing’. However, full-atomistic simulations are very calculation-consuming, that’s why simulations can only cover nano- to micro-seconds of the lifetime of a protein molecule. I believe this is the first experiment that manipulates a molecule at the same speed as it is done on a computer, hence allowing direct comparison.
If you pull the protein apart faster than a certain speed, about 100µm/s, then the molecule starts behaving very differently than it does at lower speeds. It appears that the molecule is pulled so fast that the imposed trajectory dominates over natural diffusion.
This is a fundamental biophysics technique and, as such, will allow us to examine any type of single molecule interaction – obvious targets are ligand-receptor pairs.
Go faster! Not only pulling at faster speeds, but also detecting events faster. We also hope to apply the technique to challenging bio-samples.
References
- F. Rico et al., “High-Speed Force Spectroscopy Unfolds Titin at the Velocity of Molecular Dynamics Simulations”, Science, 342 (6159), 741-743 (2013)DOI:10.1126/science.1239764
