This study demonstrates a routine screening of drugs to identify and quantify potential genotoxic compounds. In this Application Note, we used an Agilent 6545 Q-TOF LC/MS system to acquire accurate mass data of samples containing chlorhexidine as the drug substance. Agilent MassHunter Mass Profiler software was used to mine the data and compare different samples to generate a differential list of compounds. An accurate mass database search against the differential list identified 4-chloroaniline, a potential genotoxic compound. All Ions MS/MS acquisition mode was used to confirm 4-chloroaniline by MS/MS library matching, and quantify it using external standards. This workflow is suitable for batch-to-batch sample analysis for detecting and quantifying known potential genotoxic compounds. The full Application Note can be found online: tas.txp.to/0116/GenotoxicApp

Introduction
Drug substances may produce potential genotoxic compounds when they are stored for extending periods of time, or when they are stored inappropriately. Detection, identification, and quantification of genotoxic compounds is a time-consuming process. Regulatory authorities (1) require reporting of the formation of genotoxic compounds. Recent advances in software tools enables the fast and cost-effective detection of potential genotoxic compounds in complex samples. Agilent MassHunter Mass Profiler (MP) software allows the comparison of two sets of samples, and the determination of any significant differences between them. Principal component analysis (PCA) tools within MP assists the classification of compounds based on identified differentiation markers. A differentiation marker is a compound that exceeds a defined concentration, when compared to a control sample. A custom-built accurate mass database was used to identify the differences between samples. In this study, MP analysis of degraded and nondegraded chlorhexidine samples gave a list of statistically different compounds between samples. Using an Agilent ID Browser feature within the MP software, these compounds were searched with a custom database containing potential genotoxic compounds. Compounds were further confirmed using accurate mass library matching, then quantified. Figure 1 shows the workflow used in this study.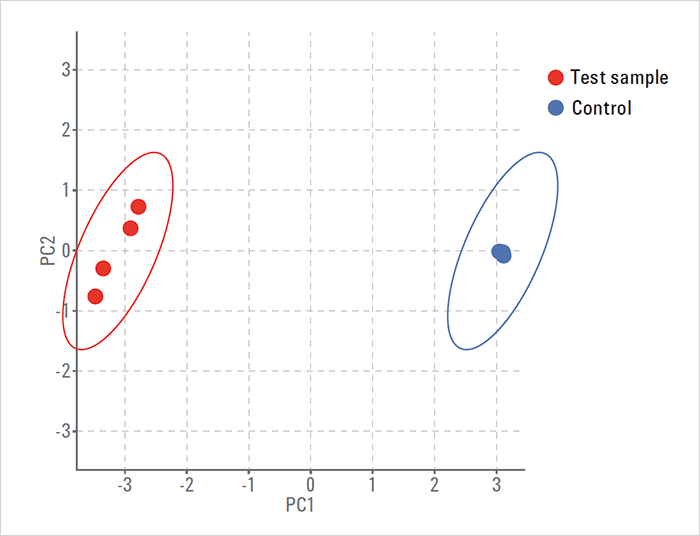
Experimental
See full Application Note for details: tas.txp.to/0116/GenotoxicAppResults and Discussion
Screening by differential analysis The data files from the LC/MS analysis of degraded and control samples were processed using recursive molecular feature extraction in Mass Profiler software. Height filters of 4,000 counts for extracted compound features, quality score 100 and >4-fold change were used for statistical analysis. A greater than 4-fold change was applied to detect those features that differed significantly from control samples. See full Application Note for more details: tas.txp.to/0116/GenotoxicApp PCA plot The PCA plot reveals that the degraded chlorhexidine samples are different and distinct from the control sample (see Figure 1). This indicates that the degraded chlorhexidine sample contains features that are different from the control group. The control groups do not show significant separation, indicating no variation (blue dots) between samples. Compound identification A customized accurate mass database and library was created using standard compounds. The database also included literature reported mass, formula, and structures of chlorhexidine impurities. Post-statistical analysis, the differential list of compounds was searched against the accurate mass database using the ID Browser feature within Mass Profiler. The results indicated the presence of a potential genotoxic, 4-chlorhexidine in the degraded samples. Feature summary of compounds See full Application Note for details: tas.txp.to/0116/GenotoxicApp Confirmation and quantification of potential genotoxic compounds A shorter data-independent acquisition method was used for the targeted confirmation and quantification of 4-chloroaniline. In data-independent acquisition (All Ions MS/MS) of drug samples, both MS and MS/MS information are generated. The fragment ions in the MS/MS spectra of the personnel data compound library (PCDL) were used to extract ion chromatograms from the high energy channel. The extracted ion chromatogram (EIC) of the precursors from the low energy channel were aligned with fragment/production EICs to obtain the coelution score. The 4-chloraniline was confirmed based on accurate mass fragment matching and coelution of the precursor and product ions. 4-Chloroaniline was found with three qualified spectra in the library MS/MS spectrum where the fragments are selected from high energy MS analysis. The selected spectra were used with the qualifier and quantifier ions for the quantification method.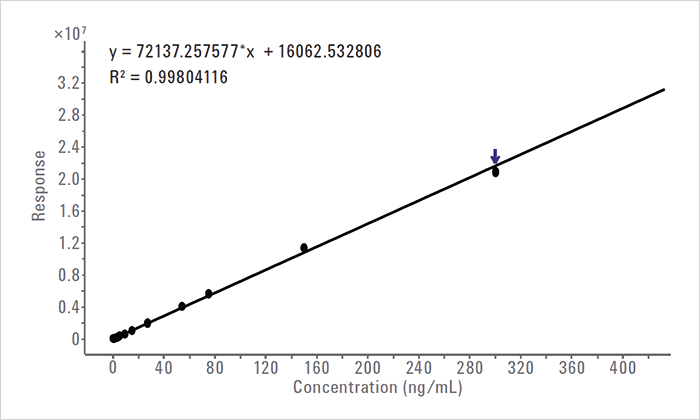
The qualifier and quantifier fragment ions, together with compound names, retention time, precursor ion, fragment ion, collision energies, and relative abundances were exported to MassHunter Quantitative Analysis software to set up a quantitative method. The most intense ion was used as a quantifier trace, while the less intense and unique fragment ions were used as qualifiers. A calibration curve with > 3 orders of magnitude was plotted from 0.1 to 300 ng/mL (see Figure 2). The 6545 was calibrated and tuned in high sensitivity mode. In addition, tuning for low mass (50–250 m/z) using Swarm autotune was enabled since some of the product ions for 4-chloroaniline were of low mass. The results of sample analysis showed an average value of 29 ng/mL in the degraded sample. Potential genotoxic compounds typically have a limit for reporting of 0.05 %. When 1 mg chlorhexidine is dissolved in 10 mL solution, a 0.05 % limit would require quantitation down to 50 ng/mL. Therefore, any assay must be capable of a lower LOQ. The method developed in this study can detect impurities present at a concentration <1 ng/mL. Conclusions This Application Note demonstrates that potentially genotoxic compounds can be screened, identified, and quantified using high resolution LC/MS. A streamlined workflow was achieved by combining All Ions MS/MS data with Agilent MassHunter Mass Profiler software (Rev. 7.0). Automated differential marker analysis revealed significant differences between sample and control sets. The workflow also included the automated detection and identification of potential genotoxic impurities as target compounds using a PCDL. The All Ions MS/MS methodology was used to generate both quantifier and qualifier ions. This enabled the quantification of the target compound. The test sample processed with this technique was determined to be at a concentration of ~29 ng/mL or 0.02 % of 4-chloraniline (assay linear range from 0.1–300 ng/mL). This workflow can be used as part of routine drug sample analysis for the identification and reporting of potentially genotoxic compounds.
References
- EMA Guidance on the limits of genotoxic impurities, EMEA/CHMP/QWP/251344/2006 http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2009/09/WC500002903.pdf

